Virulence factors determine pathogenicity
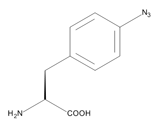
The numbers of infectious diseases caused by human pathogenic yeasts have been continuously increasing over recent years. Their high morbidity and mortality have turned them into a serious public health problem. An efficient treatment is particularly complicated by systemic mycoses, which spread throughout the whole body, or emerging resistance to antibiotics. The most prevalent causative of systemic mycoses in humans is the pathogenic yeast Candida albicans, which can elicit severe infections if the immune system of its host is suppressed by, for example, operations, chemotherapy or diseases. Candida albicans features a multitude of mechanisms which lead to pathogenicity. These mechanisms are mediated by virulence factors, proteins with various functions in the cell. Virulence factors are essential for pathogenicity and therefore appear to be a promising target for the development of therapeutics. However, this requires profound knowledge of the molecular characteristics and physiological interaction networks of cell proteins. As techniques to study protein interaction networks in vivo are scarce, especially for C. albicans, scientists at the Fraunhofer IGB developed a new method to analyze protein-protein interactions with artificial amino acids.
 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB