Focus on Infection, Inflammation and Innate Immunity
Immunomodulators for therapy
The central objective of the JRHDD project is to exploit mechanisms of innate immunity in combination with targeted drug delivery for therapy of infections as well as autoimmune and inflammatory diseases. This idea has been fostered by results already achieved in a joint project (ICON project between Fraunhofer IGB and the Hebrew University) [1, 2]. Based on these results, we aim to design immune-modulatory compounds in order to address infections and inflammatory diseases more effectively. Targeting these compounds directly to the site of infection or inflammation will support the healing process significantly. For infections, stimulating the host's owned defense mechanisms should also be effective against pathogens resistant to the current anti-invectives, enhancing “classical” medication efficiently.
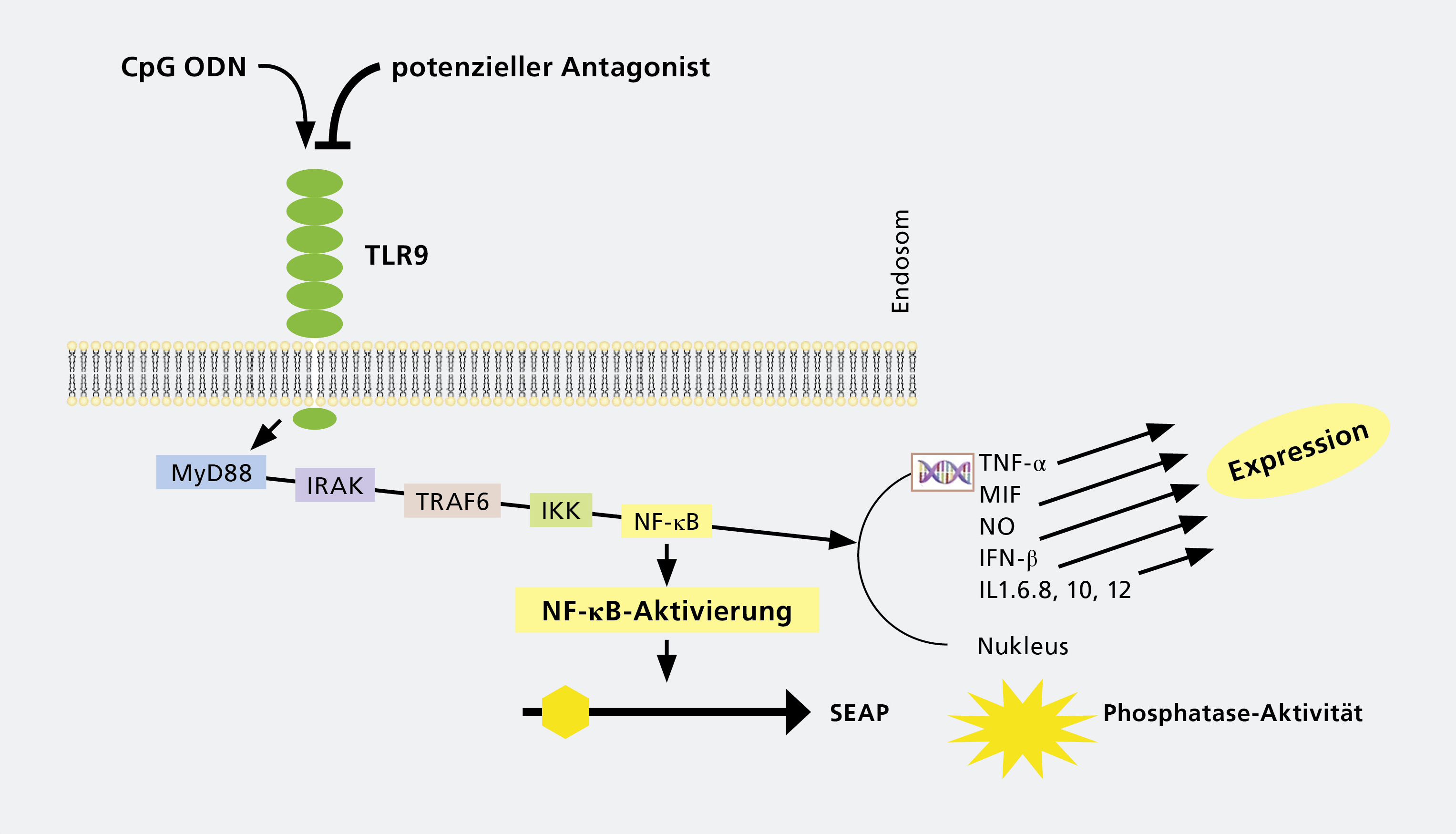
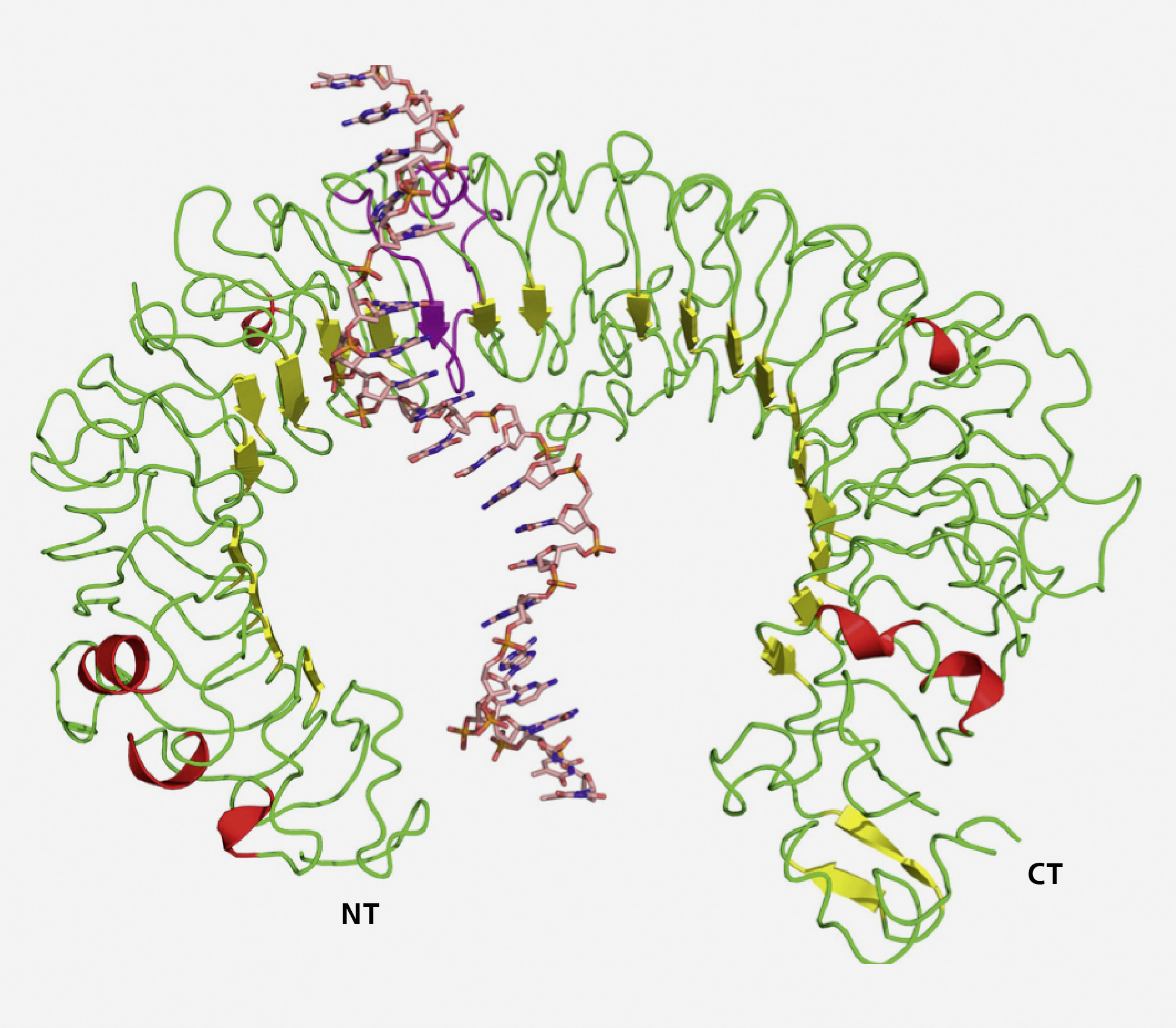
As a first target we have identified dermatological disorders, such as atopic dermatitis, psoriasis and lupus, since they show involvement of innate immunity, including toll-like-receptors (TLRs). Currently, the pipeline for mild-to-moderate psoriasis includes at least 22 investigational topical therapies in various stages of development, including IL-17 antagonizing agents, showing that immune-modulating agents have a high potential as medication. Dermatologic disorders often show a complex interplay between defects in skin barrier function, environmental impact and infectious agents as well as changes in immunity.
The second focus is to develop novel drugs and targeted nanomedicine to combat fungal infections caused by fungi such as Candida spp. and viral infections caused by Herpesviridae. Since innate immunity is an essential part of combating infectious diseases, a combination of drug development, stimulation of innate immunity and targeted formulation of the respective compounds is an ideal approach to combat infections. The first compounds with TLR-modulating activities have been introduced in clinical trials as antiviral agents, indicating the feasibility of this approach.
 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB