Proteins are macromolecules, which as a rule are formed from 20 canonical amino acids via peptide bonds in the cell. In this context, the unique amino acid sequence of each individual protein defines its biological function and its physico-chemical characteristics. Proteins are of scientific and economic importance, for example highly specific enzymes with defined catalytic properties for the synthesis of complex compounds or therapeutically applicable active substances. Currently, proteins are generated by standard recombinant technologies.
Synthetic proteins for the analysis of biomolecular interactions in vivo
Synthetic biology
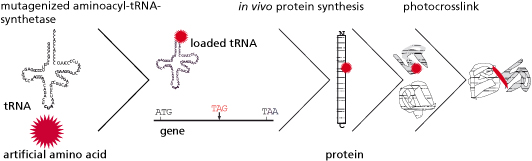
One of the most recent and promising approaches for the biosynthesis of proteins with novel physico-chemical characteristics is provided by the rapidly developing field of synthetic biology. In this context, the underlying principle allows the generation of recombinant organisms using standardized components as well as engineering principles. In addition to standard, natural amino acids, these synthetic organisms can also site-specifically incorporate artificial amino acids with novel properties into the peptide sequence in vivo. These newly added functional groups can either improve the physiological function of the protein or provide the protein with unique biochemical properties that go far beyond the natural spectrum, such as cross-linking capability, photoactivation, or the possibility of selective, posttranslational modifications.
Technology development using model system Saccharomyces cerevisiae
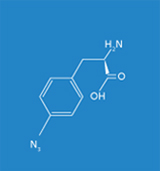
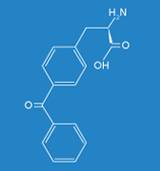
The objective of the Fraunhofer IGB is to generate synthetic proteins based on the site-specific incorporation of artificial amino acids such as azidophenylalanine or benzoylphenylalanine in vivo for studying biomolecular interactions, such as protein-DNA and protein-protein interactions, in eukaryotes under physiological conditions in vivo. For the implementation of this technology we make use of the Gal4p transcription factor in Saccharomyces cerevisiae as a model system in order to localize DNA binding sites genome-wide and to identify potential protein-protein interactions.
Principle
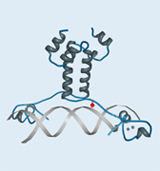
Frequently, the interactions of a protein with its environment are not stable, but only transitory. The association between the interaction partners can, for example, dissociate as soon as the biological systems being examined are disintegrated for analysis. For this reason it is essential for the analysis of protein-protein interactions that the respective biomolecular interactions are covalently linked. In this context the time of fixation should be arbitrary and specific for the interaction. To meet this requirement, we make use of an expanded genetic code (Fig. 1). In this context, instead of a natural amino acid an artificial amino acid will be incorporated into the interaction domain of the transcription factor Gal4p in vivo at a defined position during biosynthesis (Figs. 3 and 5). For this purpose, azidophenylalanine and benzoylphenylalanine, which are derived from the natural amino acid phenylalanine, are used as artificial amino acids. The additional side groups of these amino acids can then be photoactivated by UV light to form covalent links to the respective interaction partners. These protein complexes, which are very tightly bound to one another, can then be isolated, subsequently enriched, and identified by means of mass spectrometry.
Perspective
The technology for the generation of “synthetic” proteins for the analysis of biomolecular interactions has the potential to be universally applied and thus will contribute to a better understanding of complex regulatory networks in the development of diseases or to clarify metabolic pathways.
Funding
The work on the establishment of synthetic proteins was funded by the Fraunhofer-Gesellschaft within the scope of its MEF program (Research program for small and medium-sized enterprises) under the title “Procedure for the genome-wide identification of regulatory protein-DNA interactions”.
 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB