Gene expression analyses for any organism – without DNA probes
Gene expression analyses give an answer to the question, which genes are active in a cell at the moment of investigation – meaning which genes are transcribed into messenger RNA and then translated into proteins (expressed). At the Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB scientists have developed a novel high-resolution and highly sensitive process, which can be used universally for any desired non-sequenced eukaryotic organism. Therefore, the method is of great interest for research in the pharmaceutical, the agricultural and the chemical sector.
What are the differences in the active genes of a healthy and a sick person? Which genes are elsewise expressed in cancer tissue in comparison with the expression in normal tissue? Which genes make agricultural crops susceptible against fungal infections? An answer to these questions gives the differential gene expression analysis. Comparing the whole transcriptome, for example cancer tissue with healthy tissue, may help to identify so-called tumor markers - genes expressed specifically only in cancer tissue. If these genes are known, tumors can be classified and the prospect of success of distinct therapies can be estimated. In future, even specific or individualized diagnostics and therapeutics can be developed.
The toolkit of choice for genome-wide gene expression studies is the DNA microarray, also called DNA biochip. However, DNA microarray technology requires the knowledge of the complete annotated genome sequence of the organism to be studied. In addition, the preparation of specific DNA probes and microarrays is laborious and cost-intensive. In order to enable gene expression analyses beyond the well-known sequenced model organisms such as man, mouse and yeast, researchers of Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB in Stuttgart have developed a universal gene expression analysis process in cooperation with the companies GATC in Konstanz and raytest in Straubenhardt as well as with the Laboratory for Functional Genome Analysis LAFUGA at the Gene Center in Munich.
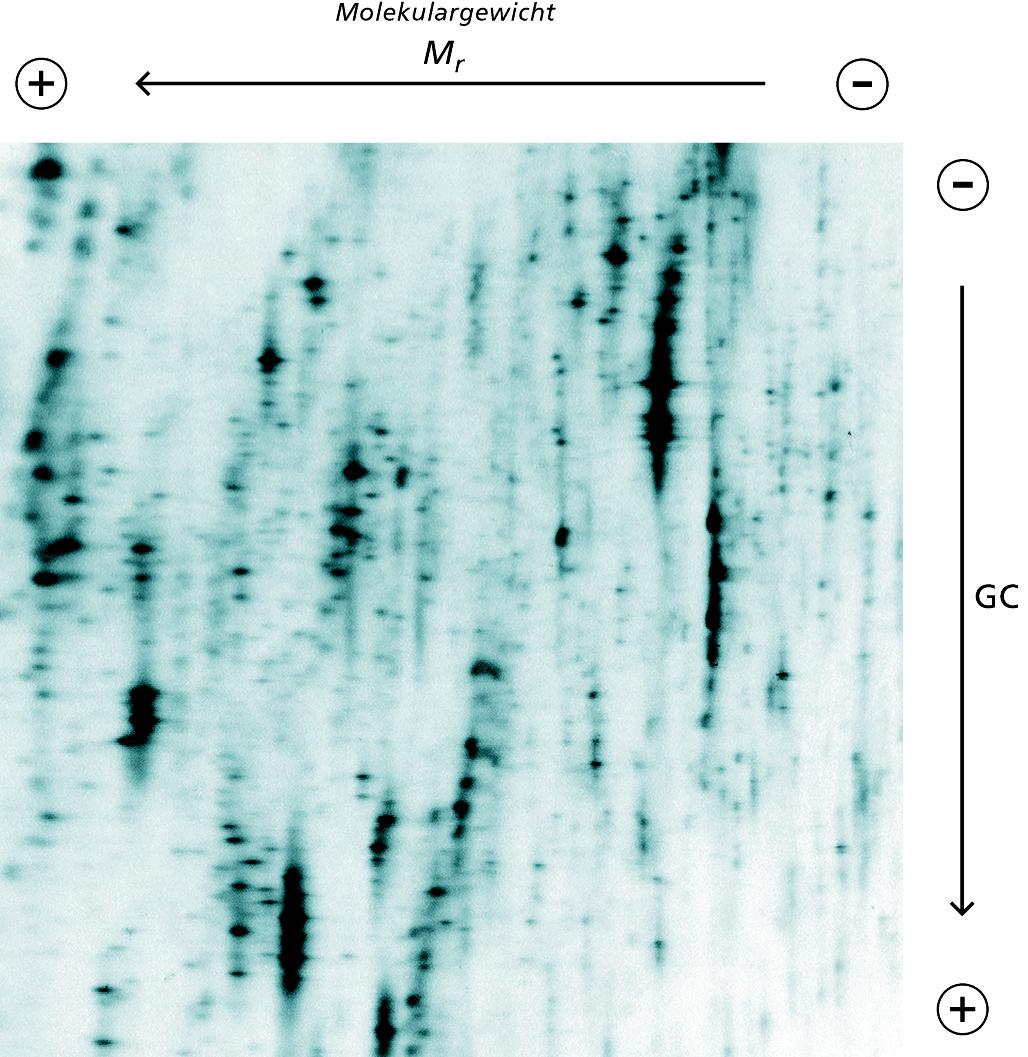
The technology is based on the high-resolution gel electrophoresis separation of complex cDNA samples. First, total RNA from the test material is isolated, transcribed into double-stranded cDNA and amplified. The cDNA is then separated by means of two-dimensional DNA gel electrophoresis. The DNA fragments are first separated according to their molecular weight, in the second dimension according to their GC content. The complex spot patterns thus obtained can be visualized using fluorescent dyes such as for example SYBR Gold. "By comparing the spot patterns of two distinct samples of the same organism, we can identify differentially expressed cDNAs and thus those genes which are differentially transcribed", explains Dr. Kai Sohn, project manager at Fraunhofer IGB.
The great advantage of the new procedure is its universal application. "We can investigate any organism within the plant and animal kingdom. We can analyze agricultural crops, which are resistent against pathogens. We can also analyze domestic animals as well as virulent fungi not sequenced so far", says Kai Sohn. The procedure is very sensitive. "For each analysis we require only 1 microgram of total RNA, whereas a DNA array typically requires 25 micrograms. Furthermore, we can identify yet unknown small transcripts, which are not detected by DNA microarrays", Sohn emphasizes further advantages of the technology filed for a patent.
The procedure is of special interest for companies and research laboratories from medicine, biotechnology and plant breeding wishing to identify molecular targets in complex model systems in order to develop specific diagnostics or to optimize crops.
 Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB
Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB